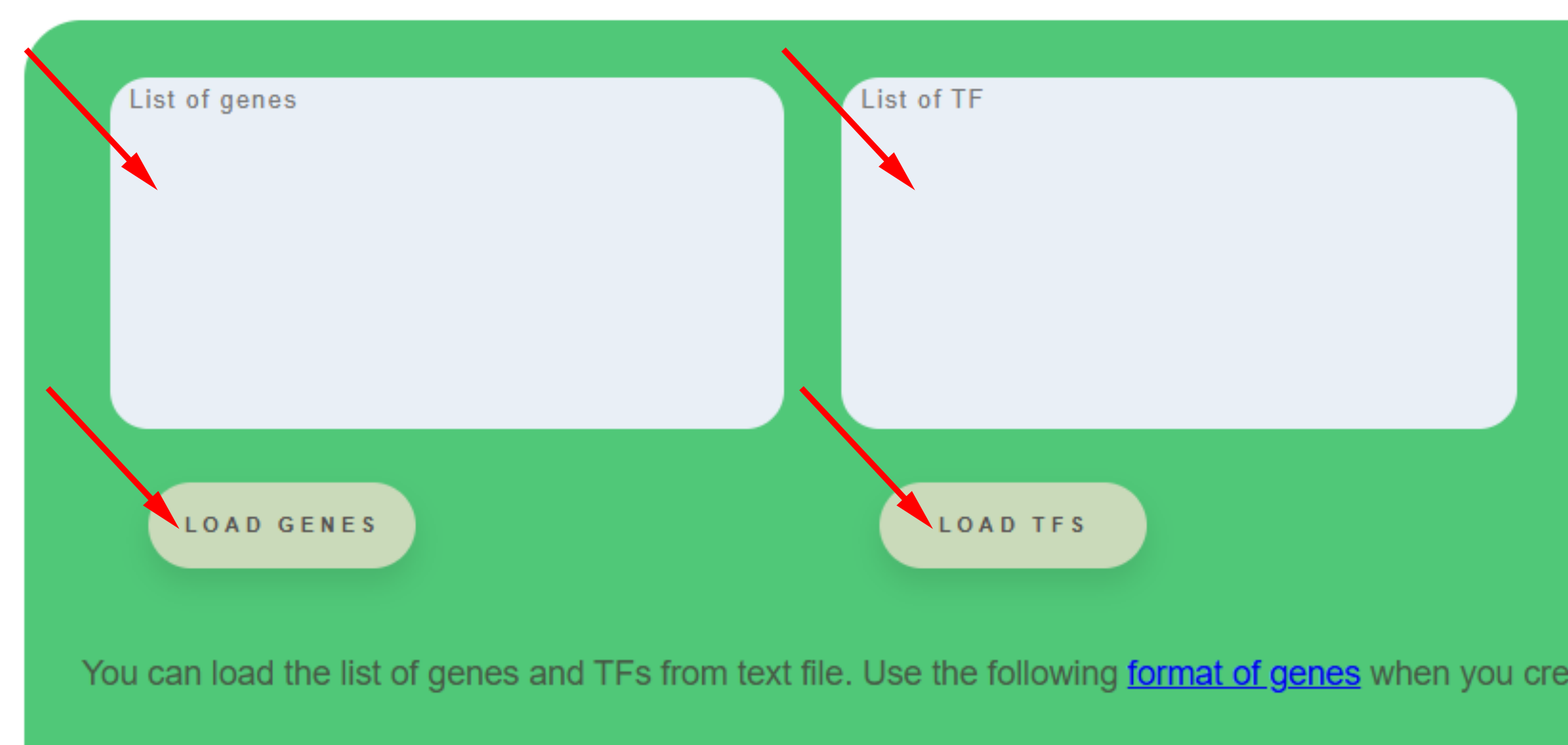
Web server CisCross-TF-targets is an application for searching amount input transcription factors (TFs) list potential regulators of the input gene list.
CisCross-TF-targets takes a list of genes and a list of TFs. A user can use any list of genes. It also could be differentially expressed genes. Publicly available gene expression databases, for example the EBI Expression Atlas (Moreno et al., 2022, https://www.ebi.ac.uk/gxa/home) is a good source of qualitative transcriptomic data. As a list of TFs a user can сhoose TFs that are known or putative transcriptional regulators of the genes as input.
The Button Load genes allow fill the field List of genes or insert gene list from the clipboard. Genes should be name with TAIR ID, for example AT1G05680. On the web page, there is the recommendation for creating a file (format of genes). The Button TFs allow fill the field List of TFs or insert TF list from the clipboard. Names of TF should be name of its genes as TAIR ID, for example AT1G36060. On the web page, there is the recommendation for creating a file (format of genes).

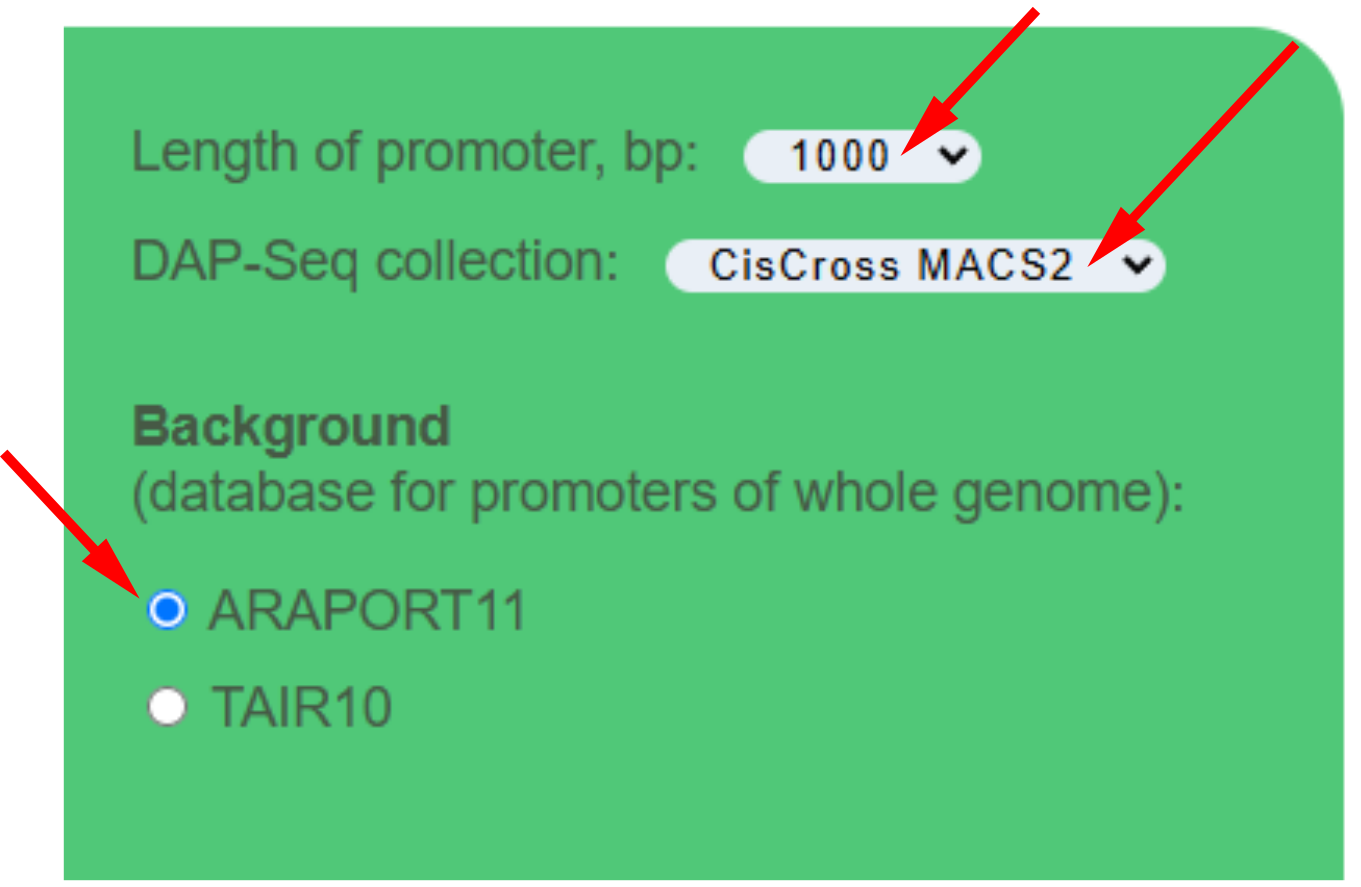
All required sections contain default values. To run the algorithm of finding targets of TF list among the input gene list push the button Submit.
To test the workflow use the button Example
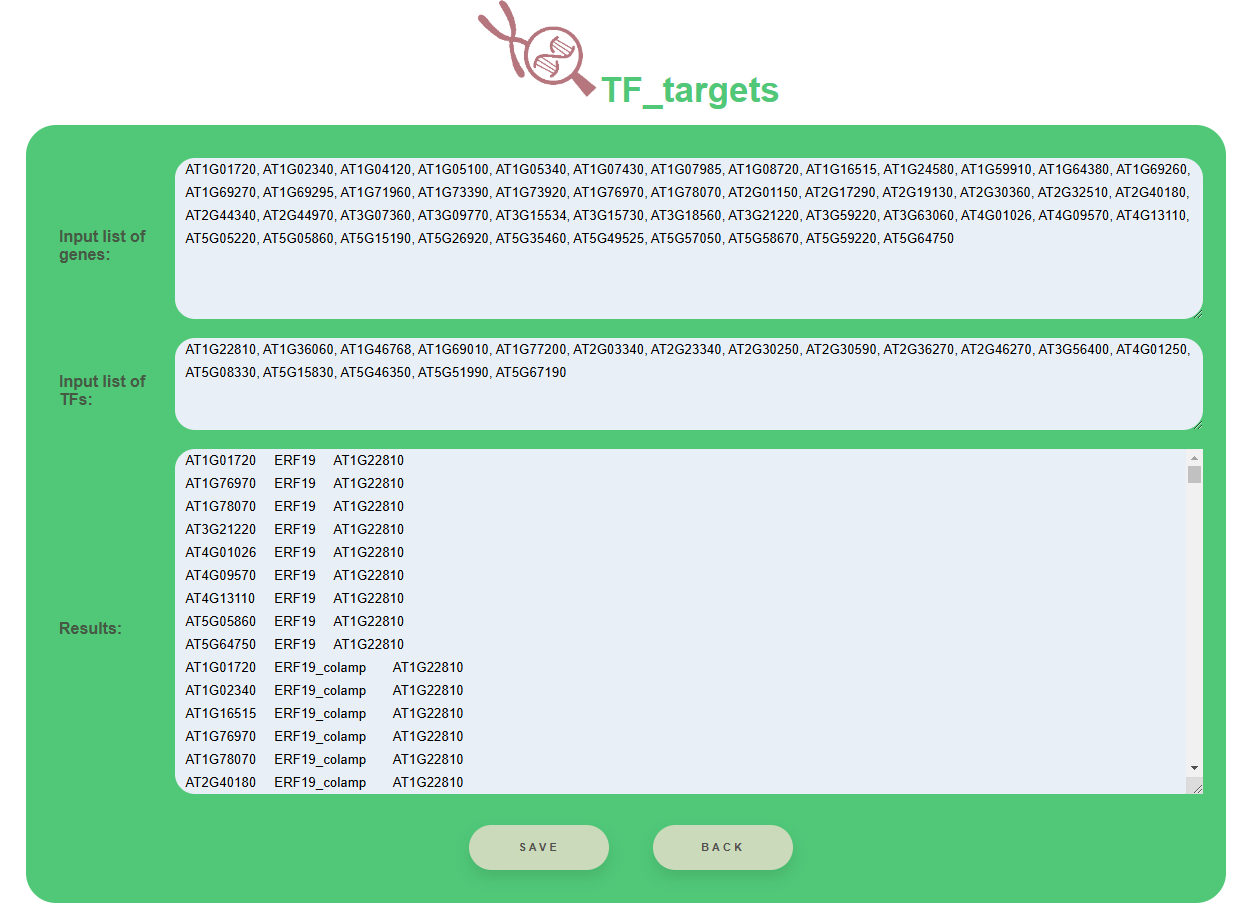
In the Results section, there are only TFs from chosen DAP-seq collection. Lists of TFs in each collection are here
Extending information about TF families assignment
To save results use the button Save below Results section.