Manual page for web service CisCross-Light
General description
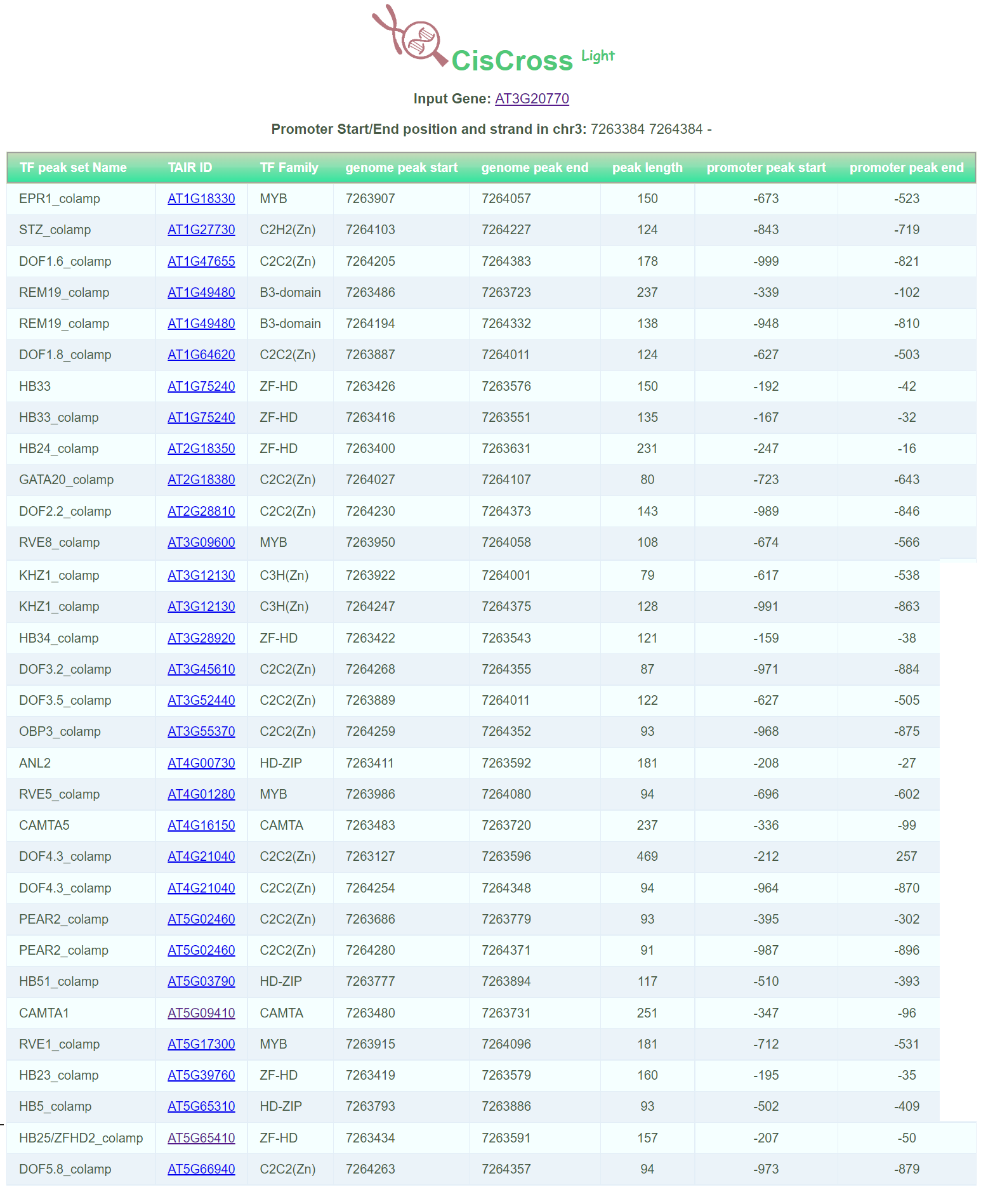
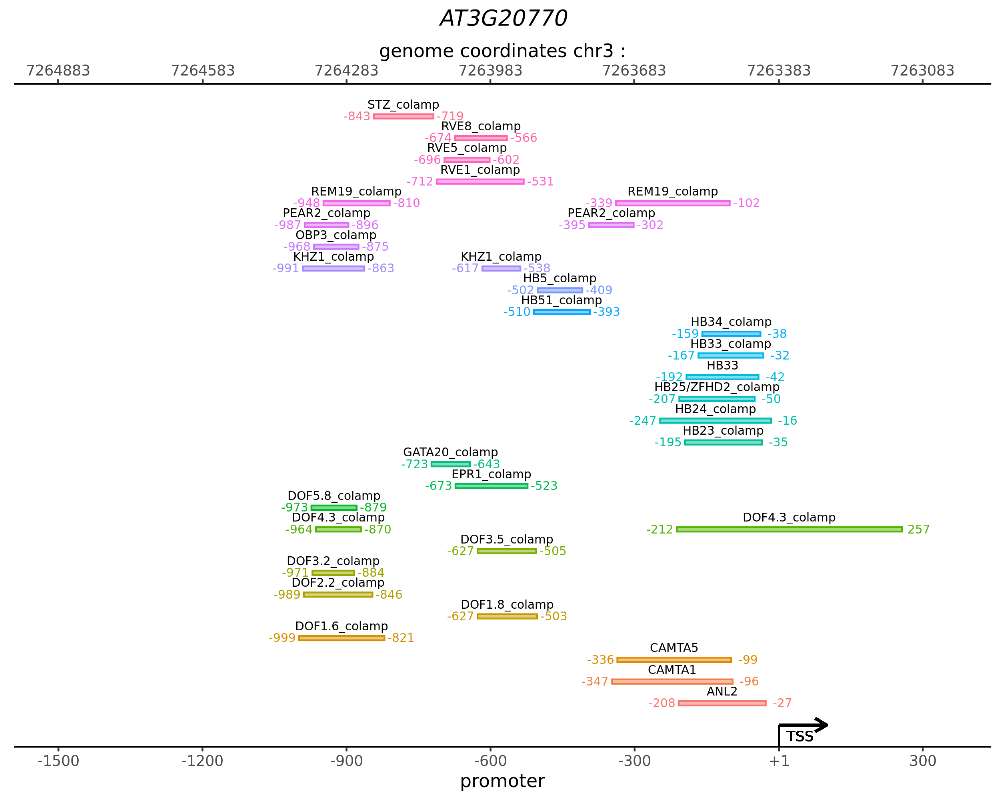
Web service CisCross-Light gives the list of DAP-Seq peaks detected in the 5’-regulatory region of the given input gene
Input data
CisCross- Light takes TAIR ID of the input gene, for example AT3G20770.
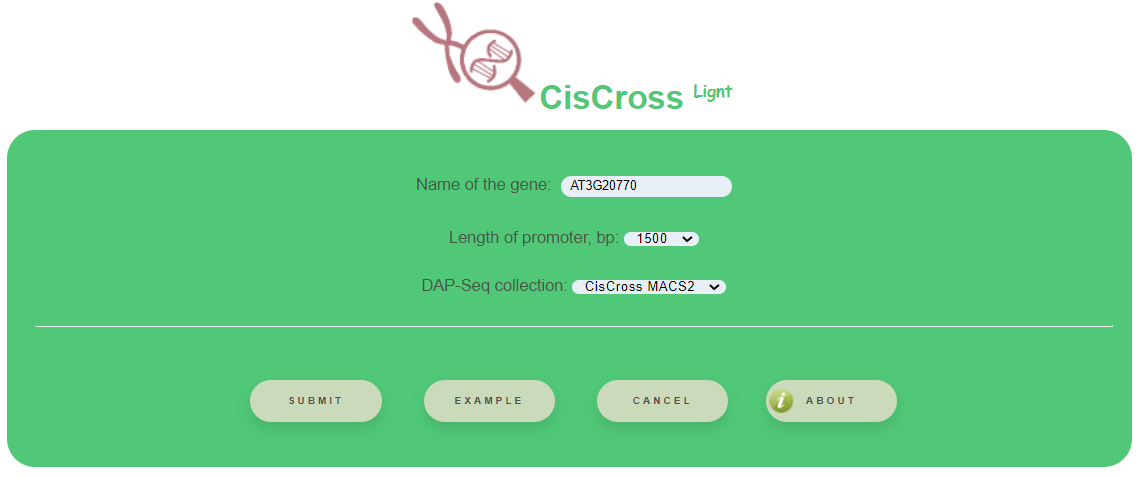
CisCross-Light requires the next selections:
- Length of promoter (bp) – whole-genome annotation of gene promoters as the regions of 500, 1000, 1500, 2000 or 2500 bp upstream to the transcription start sites
- DAP-Seq collection – the available versions for the DAP-Seq peak set collection are (1) Plant Cistrome (O'Malley et al., 2016); (2) CisCross-GEM (GEM-processed); (3) CisCross-MACS2 (MACS2-processed)
Output data
Output data of CisCross-Light represents the list of DAP-Seq peaks of TFs detected in the 5’-regulatory region of the given input gene with coordinates of the peak start and peak end.
To save results use the button Save below list of input genes.