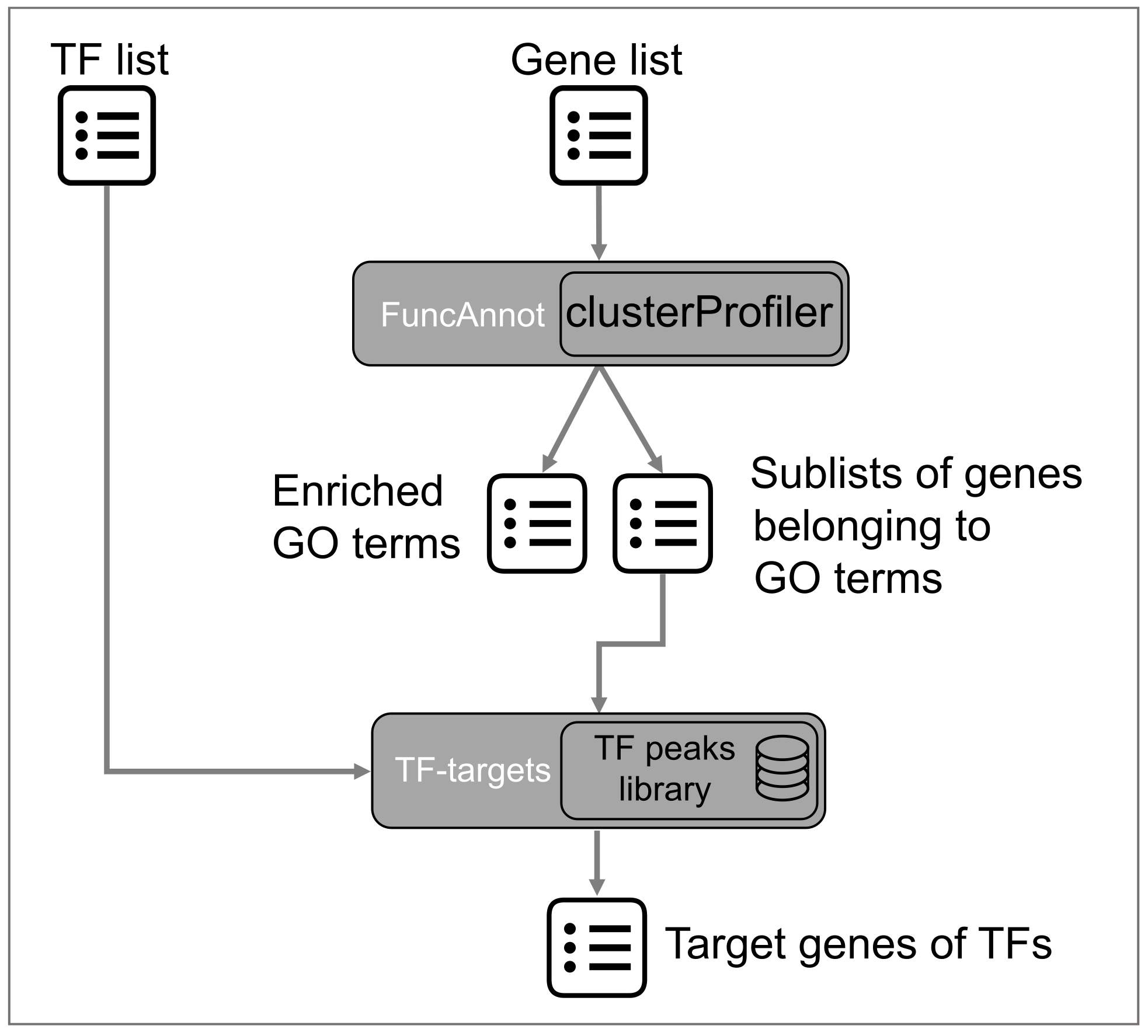
The PlantReg tool implements the reconstruction of the links between transcription factors and biological processes under their control.
The program takes a list of genes (also it could be a differentially expressed gene (DEG) list) and a list of TFs that are known or putative transcriptional regulators of these genes as input.
The FuncAnnot function performs functional annotation of the gene list using clusterProfiler R package (Yu et al., 2012; Wu et al., 2021b). The result is the information about the GO terms enriched in the gene list, as well as sublists of genes from the input annotated to the enriched GO terms.
The next step is the search of the overlaps between the binding peaks for the input TFs and 5′-regulatory regions of genes from the sublists. For this purpose, the TF-targets function, which we developed earlier as part of the CisCross-FindTFnet program (Omelyanchuk et al., 2024), is applied.
As output, a user receives a file containing enriched GO terms and their associated DEGs, evidence codes, and TFs whose binding peaks are mapped to the 5' regulatory regions of DEGs associated with enriched GO terms.ne.

In the output there are only TFs from chosen DAP-seq collection (CisCross MACS2 or GTRD MACS2). Lists of TFs in each collection are here